Biomedical Named entity recognition - Pros and cons of rule-based and deep learning methods
The final blog in our series on text-mining is a guest blog written by Shyama Saha, who specialises in Machine Learning/Text Mining at EMBL-EBI. The CINECA project aims to create a text mining tool suite to support extraction of metadata concepts from unstructured textual cohort data and description files. To create a standardised metadata representation CINECA is using Natural language processing (NLP) techniques such as entity recognition, using rule-based tools such as MetaMap, LexMapr, and Zooma. In this blog Shyama discusses the challenges of dictionary and rule-based text-mining tools, especially for entity recognition tasks, and how deep learning methods address these issues.
Named entity recognition (NER) is pivotal for many natural language processing (NLP) and knowledge acquisition tasks. Named Entities (NEs) are real-life objects that are proper names and quantities of interest. In a dictionary or rule-based NER system, a dictionary of terms/phrases or several rules are created based on the existing knowledge-base or vocabulary for an entity type. In the subsequent step, these dictionary terms are tagged in the text using a string exact match (Figure 1a) or a variation term that follows the defined rule. It is a fast, reliable and effective way to identify entities in a large volume of text, making it a popular NER method. However, it requires a significant amount of human effort to build a complete vocabulary, with target-specific rules, and it is nearly impossible to create an exhaustive list of terms or rules due to the ever-evolving nature of the human language. Secondly, novel terms or rules are introduced unceasingly in an active research domain such as the biomedical field.
For example, COVID-19 and SARS-COV2 are new additions to the disease and organism dictionaries.
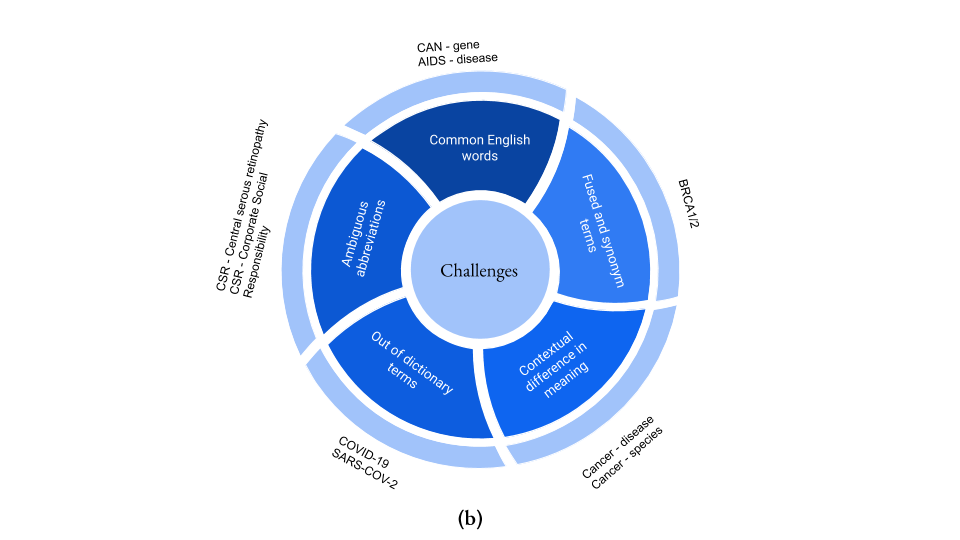
Figure 1 (a) An example of the dictionary-based entity recognition system. Due to predefined vocabulary or rules, dictionary and rule-based approaches are unable to identify ambiguous abbreviations or novel entity name variations.
Figure 1 (b) The dictionary or rule-based named entity recognition (NER) systems are popular, although there are certain challenges associated with them. This figure highlights some of these dictionary-based NER challenges.
Moreover, fused terms, term variability or novel synonyms, common English word overlapping with the entity names, and ambiguous abbreviations make development of a comprehensive dictionary even more difficult. Short gene/protein names are particularly problematic as they often overlap with the common English language words. A list of these overlapping terms is often used as a blacklist to avoid false-positive identification, although this approach results in the loss of true positive occurrences of those terms. In Figure 1b, we summarize the most common challenges of dictionary-based NER systems.
At Europe PMC, the articles are currently annotated with twenty-four entity types. Custom dictionary and rule-based annotation pipelines are used to annotate five commonly mined biological entities like gene/protein, disease, organisms, chemicals and experimental methods, and three external links including accession numbers, resources, Gene Ontology terms. Europe PMC is highly diverse, receiving sixteen other entity annotation types from its external collaborators making it one of the biggest annotated article resources. Through NER and external collaborations such as Open Targets and IntAct, we mine associations and interactions between different NEs. Moreover, Europe PMC recognises identifiers to existing archives such as BioSamples, BioStudies, BioProjects in articles and links to those data sources helping users smoothly transition from the publication to the raw data. The Europe PMC platform provides a free text search in addition to the advanced NER based query service. It allows targeted article mining through the platform front-end or programmatically via a REST API. Other text-mining research groups can leverage those annotations to carry out further research on the annotated text.
CINECA currently uses a set of text mining pipelines specialised for the different cohort (e.g., CHILD, CoLaus, etc.) and entity types (e.g., disease, medication, etc.). These approaches have been described in previous blog posts [Uncovering metadata from semi-structured cohort data, LexMapr - A rule-based text-mining tool for ontology term mapping and classification, Assigning standard descriptors to free text]. We are aiming to provide a unified portal allowing the different methodologies to be used and integrated from a single interface by the end of 2021.This enables users to query and stratify the cohort data based on sample type, disease, mutations etc. In the next section, we show how dictionary and rule-based approaches can be complemented by deep-learning approaches while dealing with a diverse and large scale unstructured data.
Deep learning approaches have gained a lot of momentum and shown a significant improvement in NER tasks in the last few years. These methods supersede dictionary-based, rule-based and the traditional machine learning NER approaches. Deep learning methods learn intricate and complex features of language using a non-linear activation function and eliminate the expensive step of feature engineering by domain experts required by the traditional machine learning methods. BERT (Bidirectional Encoder Representation from Transfer) (Devlin et al. 2019) is one of the most popular transformer models that produce a state-of-the-art result for NER task (Figure 2). BioBERT (Lee et al.) is the biomedical version of the BERT language model for the biomedical text and widely used by the biomedical text-mining domain experts for NER, question answering and summarization tasks. Although BioBERT solves several disadvantages of the traditional dictionary and rule-based approach, it has its challenges. Firstly, it is computationally expensive to train these models from scratch. Although pre-trained models are freely available and only the task-specific fine-tuning is necessary to use bioBERT, depending on the size of the training corpus it may still require several GPU-days.
Figure 2: Schematic of the BERT model (Devlin et al. 2019). The pre-training phase creates the language model and the fine-tuning provides the task-specific parameter tuning.
Deep learning methods also work as a black box making it difficult to explain the output. These are the technical aspects of a deep-learning-based NER system. There are challenges regarding the identified NERs. We have generated a gold-standard NER dataset for Europe PMC and trained a BioBERT model to identify targets (gene/protein), diseases and organisms. The model shows significant improvement in NER accuracy with maximum gain for targets in comparison to the dictionary-based approach. Our analysis shows the BioBERT model identifies a large number of novel entities. Without time-consuming, expensive, and rigorous effort, it is impossible to assess and scale the performance of the model for the full article set and determine how many of the novel terms are truly novel.
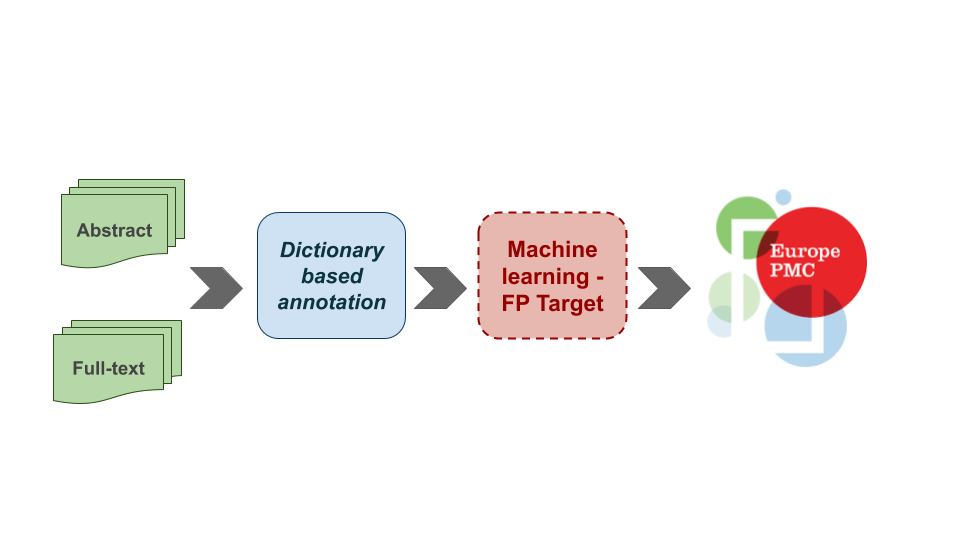
Figure 3: A hybrid approach to identify named entities in the Europe PMC articles. It is a fast and reliable way to benefit from the deep-learning approach. We are restricting false positive identifications of a dictionary-based approach in addition to the unverified novel entities from the machine learning method through the hybrid approach.
Therefore, we have taken a hybrid approach to benefit from both the dictionary-based and deep learning NER systems. We are currently using the BioBERT model to remove false-positive target terms. The hybrid approach (Figure 3) seems to be a good compromise while devising a reliable way to confirm novel entities reported by the machine learning methods. Further analysis shows the machine learning method often removes true positive terms affecting the overall performance. A post-processing layer based on the length of the entities and the removal frequency of a term across the entire article set will significantly reduce the removal of the true positive terms and improve entity identification. A similar approach would be used in CINECA.
As CINECA progresses towards producing a text-mining tool to support extraction of metadata concepts from unstructured textual cohort data and description files,we will explore ways to synergise between EuropePMC and CINECA to deploy and evaluate the impact of hybrid approaches for NER. Those will complement existing dictionary and rule-based approaches, ultimately enabling linking between literature information and standardised cohort data. Together, those will improve discoverability and reuse of CINECA knowledge.