Addressing Challenge 1: Federated data discovery.
We need to develop standardised methods and portals for federated search and discovery of relevant human data. Current development centres on using GA4GH Beacon for discovery queries, the GA4GH Search standard for extended queries, and Service-Registry for the underlying service catalogue.
CINECA aims to accelerate disease research and improve health by facilitating transcontinental human data exchange, empowering researchers to analyse data across cohorts. A key first step is to have a platform that enables ‘discovery queries’ across the cohorts so that researchers can discover relevant samples, patient data, and cohorts of interest for their particular research questions.
Uniform Discovery Queries
Are there data/samples satisfying criteria X?
Follow up with cohorts, run workflows…
Fig 1: Beacon Network - A search engine across the world's beacons, enabling global discovery of genetic mutations (https://beacon-network.org/).
Once discovered, simple interactive analyses on those subsets of data should be enabled, so that researchers can ask questions such as "how many of these patients who had treatment ‘X’ had outcome ‘Y’?”; we refer to these as ‘Extended queries’.
Uniform Analysis (“Extended”) Queries
Is variant X correlated w/ outcome Y?
What is the distribution of outcomes for disease Z?
Drive portals, answer simple questions…
For long-running analyses on these discovered data subsets, researchers must be able to hand off these subsets to other services for secure data access (e.g. AAI - Work Package 2, secure data streaming - Work Package 4) which allow computationally intensive workflows to run on the data subsets; we refer to this as Handoff.
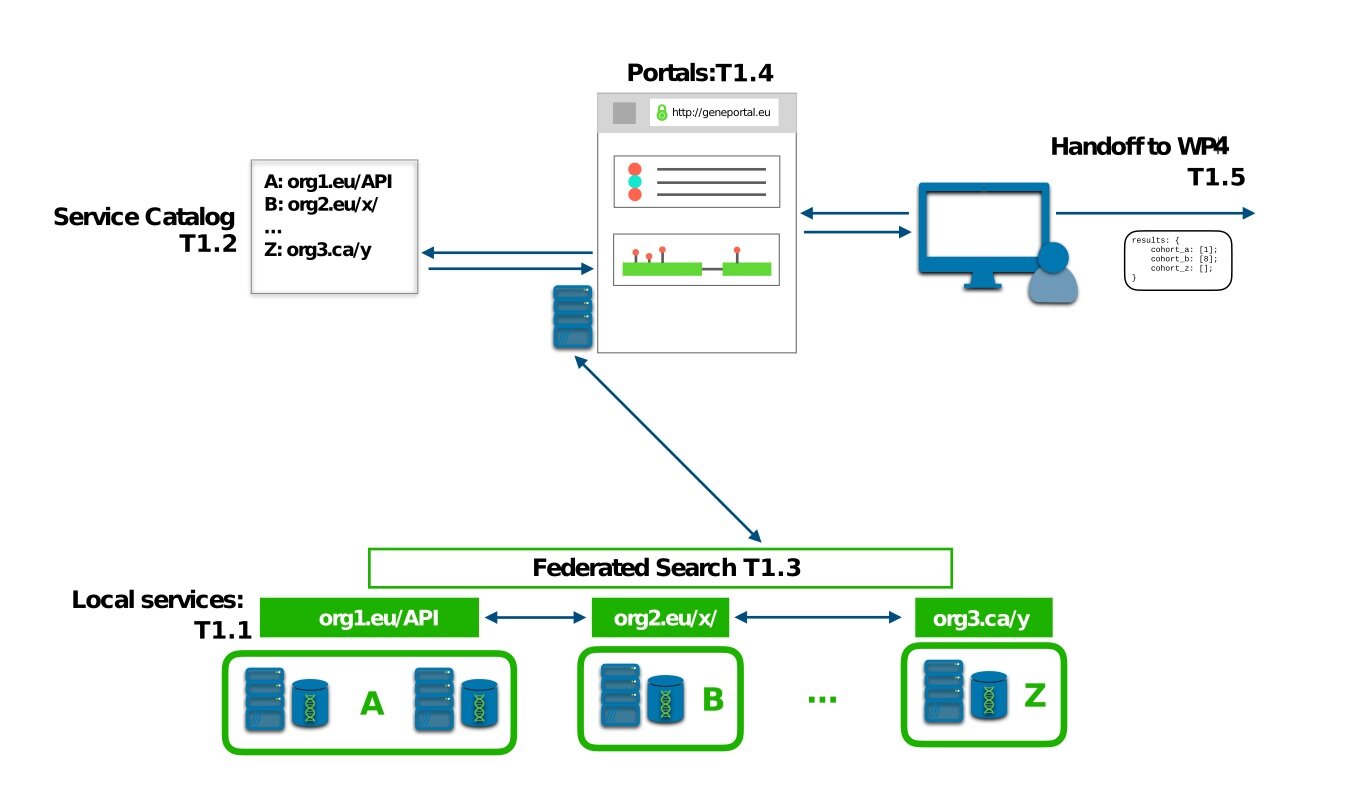
And finally, underlying the ability to connect these queries across all of the cohorts, datasets, and services provided by CINECA sites and cohorts, there must be a catalogue of available datasets and services available to the platform, which can be queried; we refer to this as the Service Catalogue.
As part of Deliverable 1.1 (June 2020), we implemented and demonstrated the final piece of the CINECA WP1 services, the Service Catalogue, which takes previous work on the GA4GH service-registry standard and extends it by providing a catalogue not only of services available, but the cohort datasets accessible through those services, using WP3’s cohort-level data standards.
Fig 2: How the pieces our team are building fit together - (1) Basic and extended query services are being implemented at the sites; (2) The portal will appear later, providing results directly to researchers; (3) Also providing results to the services of other teams for long-running analyses; (4) This enables the federated search across cohorts, including the portal; (5) This is the service registry we are developing.
By working with the standard-setting Global Alliance for Genomics and Health (ga4gh.org) and their family of existing and emerging standards, we are ensuring that our work will form part of the global network for the discovery of human cohort data resources. WP1 has implemented and deployed these standards for queries across multiple sites and several cohort and synthetic cohort datasets. In doing so, the WP1 team has developed extensions to two GA4GH standards (Beacon and Service Registry), necessary to our cohorts’ use-case and will be proposing these extensions to be incorporated back into the standards, resulting in improved standards for the community.
Work Package Leads
Jonathan Dursi, The Hospital for Sick Children, Canada
Jordi Rambla de Argila, Fundacio Centre de Regulacio Genomica, Spain