The Data Use Ontology to streamline responsible access to human biomedical datasets
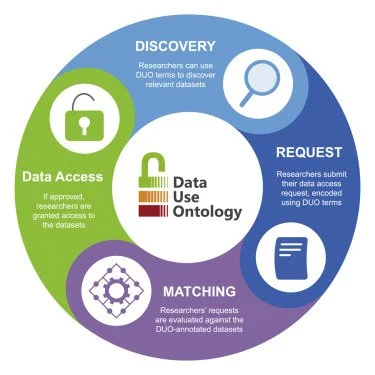
The GA4GH Data Use Ontology (DUO) provides unambiguous, machine-readable standard language for consent forms and the data sharing policies they represent. Lawson et al. describe the DUO standard and implementations throughout the data access workflow to expedite data access while maintaining or improving compliant processes.
https://doi.org/10.1016/j.xgen.2021.100028
Read More
GA4GH Passport standard for digital identity and access permissions
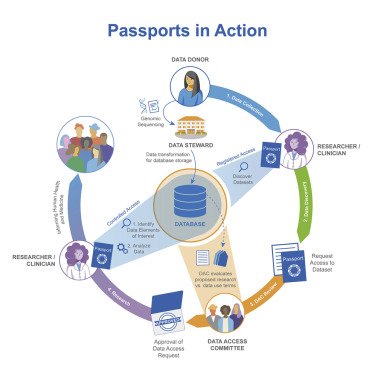
Voisin et al. report the GA4GH Passport, a new international standard to encode machine-readable data access permissions for individual users. Passports are used as part of a federated data regulatory process to authenticate and authorize data users in managing access to human biomedical datasets and have been successfully implemented in international research programs and data infrastructures.
https://doi.org/10.1016/j.xgen.2021.100030
Read More
CanDIG: Federated network across Canada for multi-omic and health data discovery and analysis
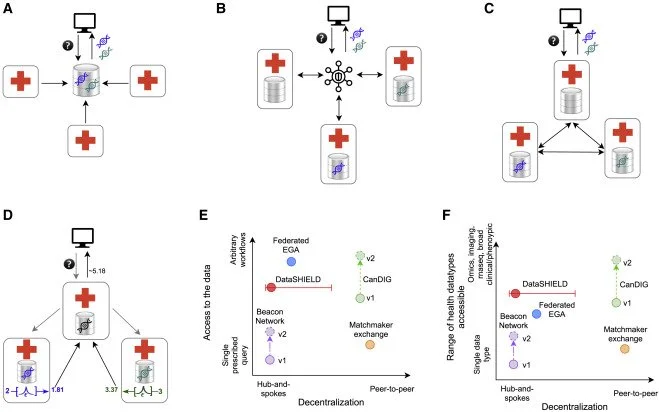
Dursi et al. describe setting up a genomics and biomedical data federation across Canada’s provincial regulatory boundaries and the drivers behind their governance and technical decisions. They guide on how to implement Global Alliance for Genomics and Health (GA4GH) standards to aid in building a federated data platform.
https://doi.org/10.1016/j.xgen.2021.100033
Read More
International federation of genomic medicine databases using GA4GH standards
Adrian Thorogood, Heidi L. Rehm, Peter Goodhand, Angela J.H. Page, Yann Joly, Michael Baudis, Jordi Rambla, Arcadi Navarro, Tommi H. Nyronen, Mikael Linden, Edward S. Dove, Marc Fiume, Michael Brudno, Melissa S. Cline, Ewan Birney
Thorogood et al. provide a guide to federated approaches to data sharing, which aim to connect independent, secure genomic medicine databases through common standards, enabling users to derive insights across multiple databases. The authors argue that a federated approach is feasible and necessary to connect national genomics initiatives into a global network to advance precision medicine.
Read More
GA4GH: International policies and standards for data sharing across genomic research and healthcare
Heidi L. Rehm, Angela J.H. Page, Mélanie Courtot, Jonathan Dursi, Lauren A. Fromont, Thomas M. Keane, Mikael Linden, Isuru Udara Liyanage, Nicola Mulder, Jordi Rambla, Gary I. Saunders, et al.
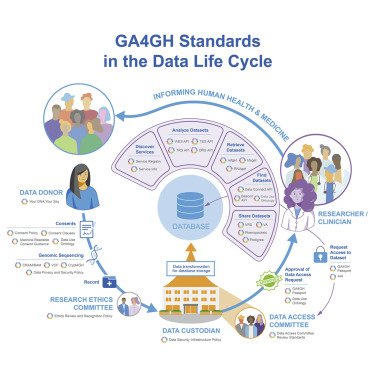
Rehm et al. describe the Global Alliance for Genomics and Health (GA4GH), which develops technical standards and policy frameworks to enable responsible international human genomic and biomedical data sharing. Broad international participation in building, adopting, and deploying these standards is necessary to bridge research and healthcare and is critical to making the best use of genomic data to inform advances in medicine and human health.
Read More
NanoGalaxy: Nanopore long-read sequencing data analysis in Galaxy
Willem de Koning, Milad Miladi, Saskia Hiltemann, Astrid Heikema, John P Hays, Stephan Flemming, Marius van den Beek, Dana A Mustafa, Rolf Backofen, Björn Grüning, Andrew P Stubbs
In this paper, de Koning, et al describe “NanoGalaxy", a Galaxy-based toolkit for analysing long-read sequencing data, suitable for diverse applications, including de novo genome assembly from genomic, metagenomic, and plasmid sequence reads.Read the full publication here.
https://doi.org/10.1093/gigascience/giaa105
Read More
Galactic Circos: User-friendly Circos plots within the Galaxy platform
Helena Rasche, Saskia Hiltemann
Rasche and Hiltemann have developed a Galaxy wrapper for Circos enabling the creation of publication-ready Circos plots using only a web browser. This version of Circos is available as an open-source installable application from the Galaxy ToolShed. Read the full publication here.
https://doi.org/10.1093/gigascience/giaa065
Read More