Authors: Nicola Mulder (UCT), Mamana Mbiyavanga (UCT), Saskia Hiltemann (EMC), Vera Matser (EMBL-EBI), Marta Lloret Llinares (EMBL-EBI)
In this deliverable document, we report on the activities in task 6.4 - Training Programme, describe the CINECA training activities that took place in months 25-36 of the project and provide the Training Plan for the final year.
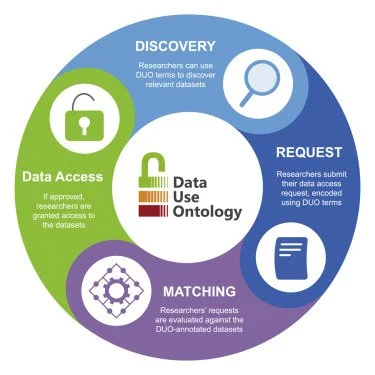
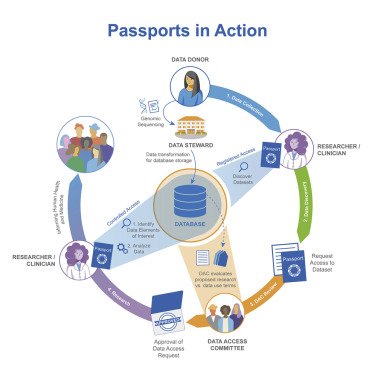
The CINECA training programme aims to train people within the CINECA consortium as well as external users. Different approaches have been employed, including face-to-face and online courses, hackathons, training videos and staff exchanges. While we waited for CINECA products to be completed, many of the training efforts for the year again focused on internal learning opportunities and knowledge exchanges, but some externally facing events were held to disseminate outputs. All the training and outreach events continued to be heavily impacted by COVID-19, which removed our ability to hold face-to-face workshops and staff exchanges. The staff exchanges were suspended and replaced with virtual meet-ups.
https://doi.org/10.5281/zenodo.5795482