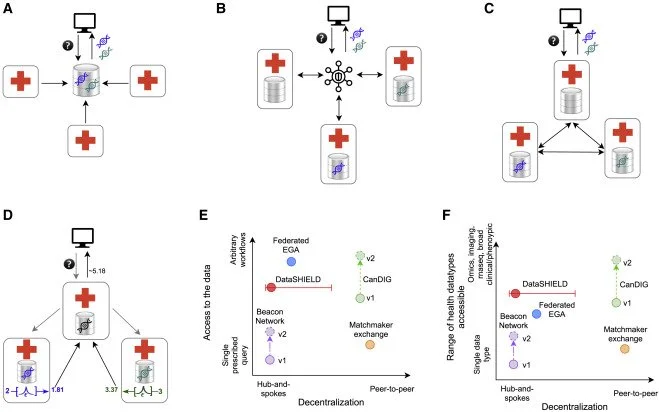
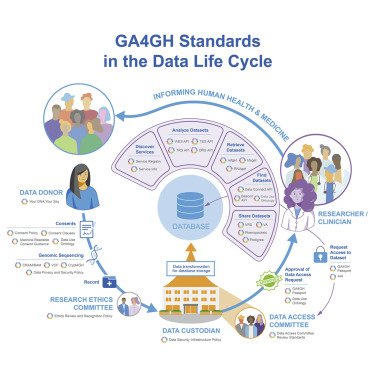
Authors - Melanie Courtot (EMBL-EBI), Isuru Liyanage (EMBL-EBI)
To support human cohort genomic and other omic data discovery and analysis across jurisdictions, basic data such as cohort participants’ demographic data, diseases, medication etc. (termed “minimal metadata”) needs to be harmonised. Individual cohorts are constrained by size, ancestral origins, and geographic boundaries that limit the subgroups, exposures, outcomes, and interactions which can be examined. Combining data across large cohorts to address questions none of them can answer alone enhances the value of each and leverages the enormous investments already made in them to address pressing questions in global health. By capturing genomic, epidemiological, clinical and environmental data from genetically and environmentally diverse populations, including populations that are traditionally under-represented, we will be able to capture novel factors associated with health and disease that are applicable to both individuals and communities globally.
We provide best practices for cohort metadata harmonisation, using the semantic platform we deployed in the cloud to enable cohort owners to map their data and harmonise against the GECKO (GEnomics Cohorts Knowledge Ontology) we developed. GECKO is derived from the CINECA minimal metadata model of the basic set of attributes that should be recorded with all cohorts and is critical to aid initial querying across jurisdictions for suitable dataset discovery. We describe how this minimal metadata model was formalised using modern semantic standards, making it interoperable with external efforts and machine readable. Furthermore, we present how those practices were successfully used at scale, both within CINECA for data discovery in WP1 and in the synthetic datasets constructed by WP3, and outside of CINECA such as in the International HundredK+ Cohorts Consortium (IHCC) and the Davos Alzheimer’s Collaborative (DAC). Finally, we highlight ongoing work for alignment with other efforts in the community and future opportunities.
https://doi.org/10.5281/zenodo.5055308
Read More