This video describes how public health genomics has played a key role in international responses to the COVID-19 pandemic, and how data standards are being used to harmonize data across jurisdictions for Canadian COVID-19 surveillance and outbreak investigations. The video is aimed at anyone interested in data standardization and/or the ontology approach (i.e. public, end users). No prerequisite knowledge is required, but viewers may also find our previous videos useful. This video is part of the CINECA online training series, where you can learn about concepts and tools relevant to federated analysis of cohort data.
Read MoreAuthors - Vivian Jin, Fiona Brinkman (SFU)
To support human cohort genomic and other “omic” data discovery and analysis across jurisdictions, basic data such as cohort participant age, sex, etc needs to be harmonised. Developing a key “minimal metadata model” of these basic attributes which should be recorded with all cohorts is critical to aid initial querying across jurisdictions for suitable dataset discovery. We describe here the creation of a minimal metadata model, the specific methods used to create the minimal metadata model, and this model’s utility and impact.
A first version of the metadata model was built based on a review of Maelstrom research data standards and a manual survey of cohort data dictionaries, which identified and incorporated overlapping core variables across CINECA cohorts. The model was then converted to Genomics Cohorts Knowledge Ontology (GECKO) format and further expanded with additional terms. The minimal metadata model is being made broadly available to aid any project or projects, including those outside of CINECA interested in facilitating cross-jurisdictional data discovery and analysis.
https://doi.org/10.5281/zenodo.4575460
The final blog in our series on text-mining is a guest blog written by Shyama Saha, who specialises in Machine Learning/Text Mining at EMBL-EBI. The CINECA project aims to create a text mining tool suite to support extraction of metadata concepts from unstructured textual cohort data and description files. To create a standardised metadata representation CINECA is using Natural language processing (NLP) techniques such as entity recognition, using rule-based tools such as MetaMap, LexMapr, and Zooma. In this blog Shyama discusses the challenges of dictionary and rule-based text-mining tools, especially for entity recognition tasks, and how deep learning methods address these issues.
Read MoreThis post is part of a series on a text-mining pipeline being developed by CINECA in Work Package 3. In previous instalments, first, Zooma and Curami pipelines were explained in "Uncovering metadata from semi-structured cohort data". Then, LexMapr was introduced in "LexMapr - A rule-based text-mining tool for ontology term mapping and classification". In this third instalment we are going to explain the normalisation pipeline developed at SIB/HES-SO.
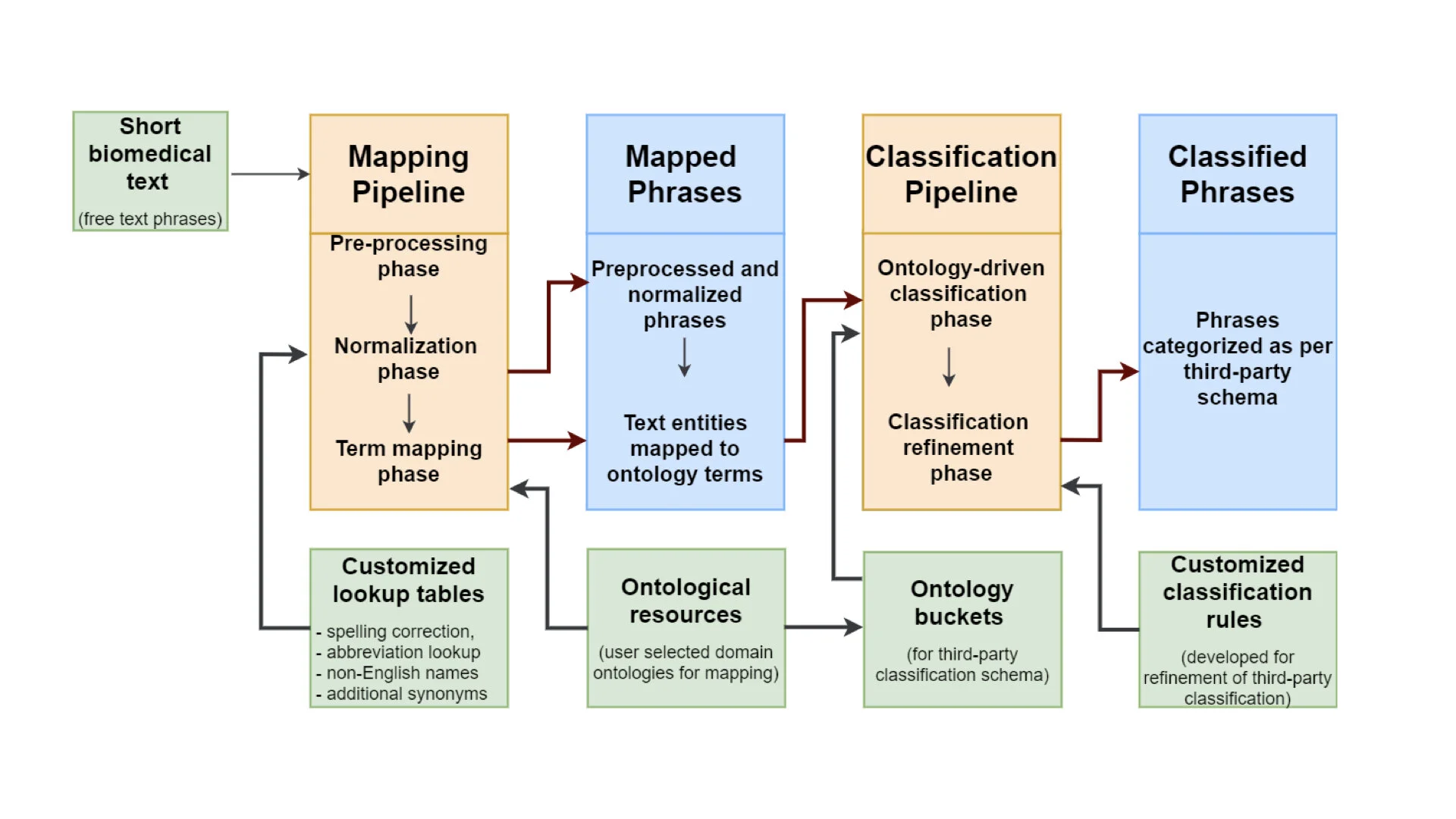
Read MoreThe initial focus of LexMapr development has been on providing a text-mining tool to clean up the short free-text biosample metadata that contained inconsistent punctuation, abbreviations and typos, and to map the identified entities to standard terms from ontologies. This blog is the second in a series on text-mining in CINECA. For the previous instalment "Uncovering metadata from semi-structured cohort data" please click here.
Read MoreHarmonisation of attributes across different cohorts is very challenging and labour intensive, but critical to leverage the collective potential of the data. The CINECA text mining group aims to provide common tools and methods to extract additional metadata from structured and semi-structured fields in cohorts’ data.
Read More